It was in 1982 when Nadrian Seeman first conceptualized the idea of using DNA as a structural element for arranging proteins in a lattice[1]. The introduction of DNA Origami technique by Paul Rothemund in 2006, further established the usage of DNA as a structural material[2]. The programmable self assmbly of DNA along with advanced in-silico design strategies, have realized the opportunity to engineer highly structured synthetic materials and systems in a bottom up fashion.[3] Since then, a wide range of DNA nanostructures of custom 2D and 3D formations have been reported for potential applications in chemical, biological and physical sciences. [4, 5, 6, 7, 8]
DNA Origami in the Nanorover, acts as an assembly platform to connect the Dyna Bead and the liposome, signifying the culmination of material science and synthetic biology via DNA Nanotechnology. A DNA origami tube comprising of a bundle of 6 DNA helices (6HB) was used as the model of choice for this purpose. Well established structural studies of these 6 helix bundles, make them an ideal choice as the constitutional framework of the Nanorover.
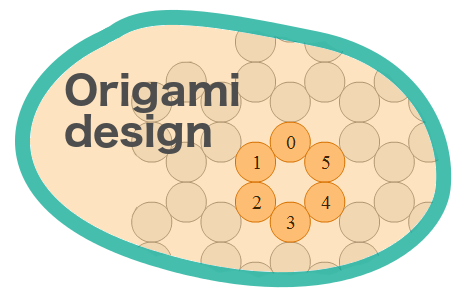
Autodesk Nanodesign software was used to further model the design from Step 1. Eight attachment sites also called "handle sequences", protruding radially outward were designed for further functional modifications. The ability to analyze the melting point profiles for individual helices was particularly useful for fine tuning the structural aspects of the 6 helix bundles.
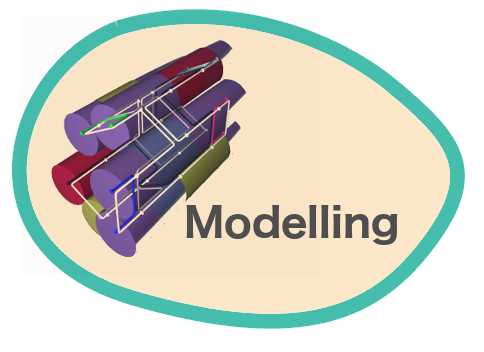
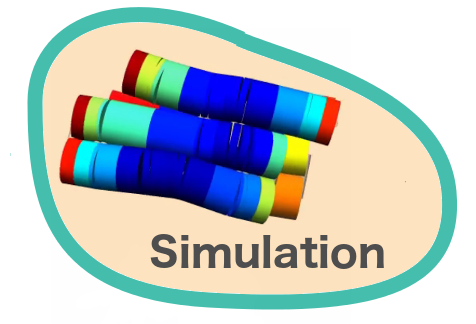
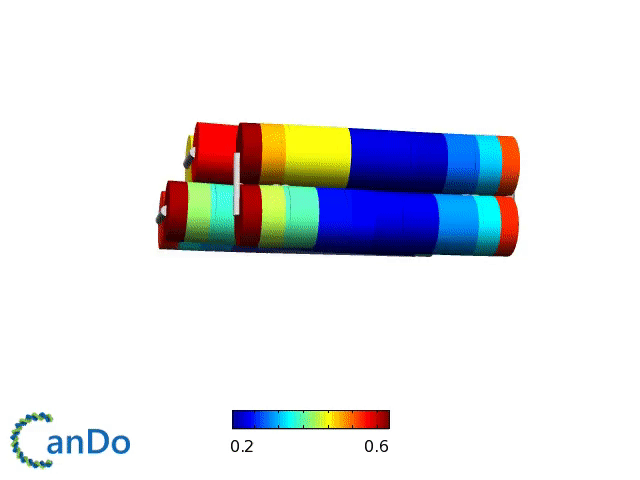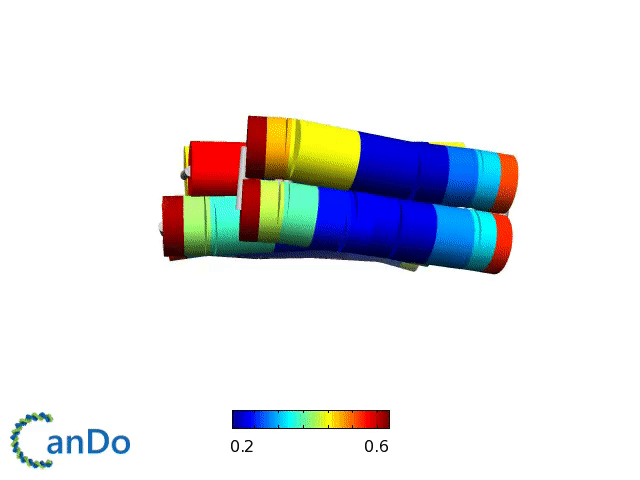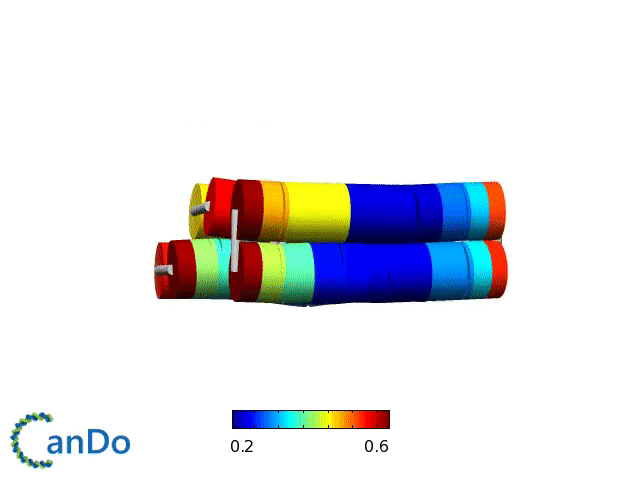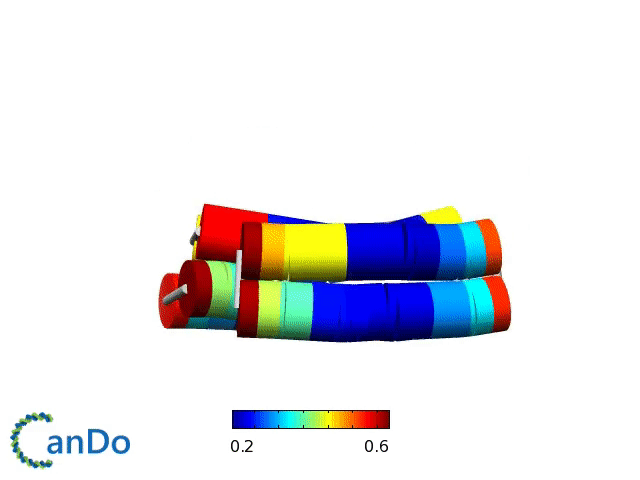
Verification of the robustness of 6 helix bundles was obtained by running CanDo solid-body simulations. CanDo simulation results as depicted in the adjacent figures confirmed that the origami designs would be able to provide global linearity due to there stiffness.

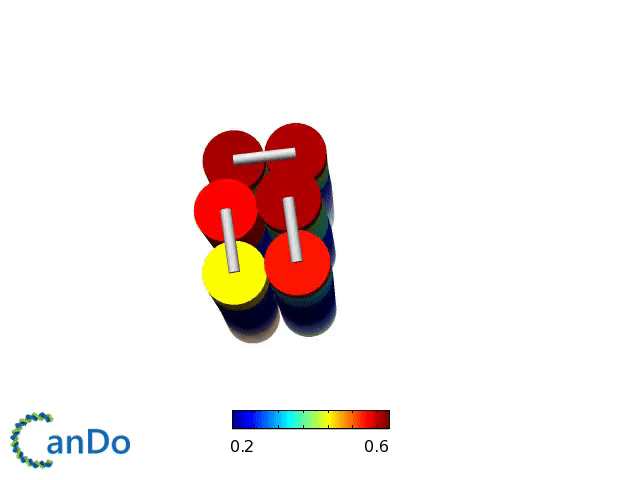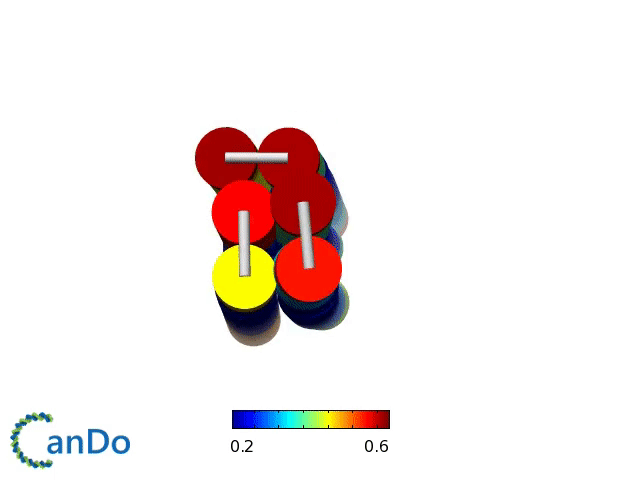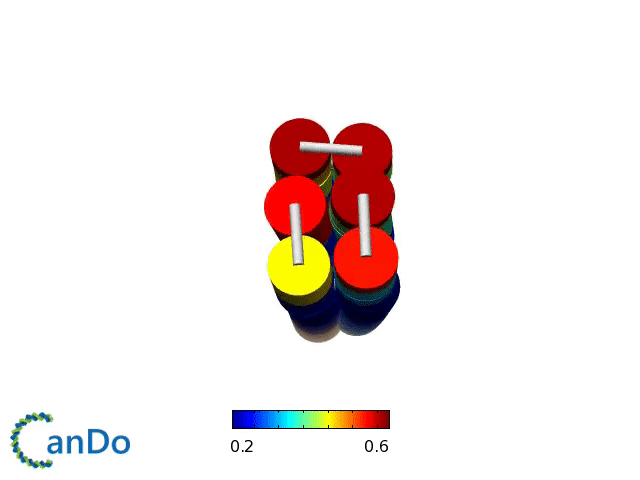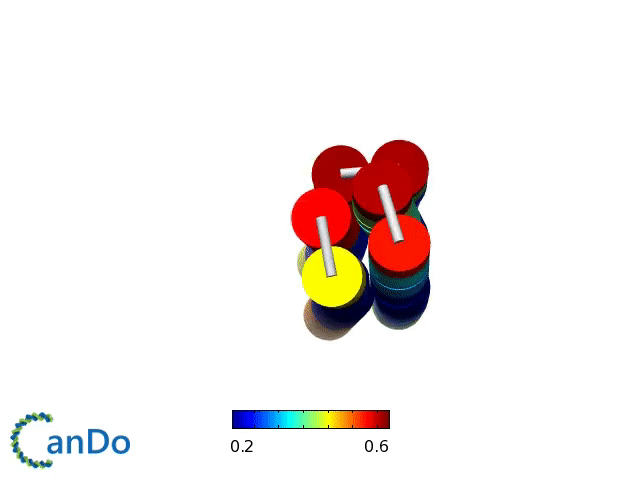
The origami was constructed using the scaffold with 8064 nucleotides in a one pot annealing reaction. The assembly was completed by using short synthetic staples strands which make up the core structure and also hybridize with binding sequences for the attachment of biotinylated liposomes as well as functionalised Dynabeads.
The folded 6HB were modified by attaching streptavidinylated ssDNA to serve as multiple attachment points for biotinylated liposomes. Furthermore, an additional loop in the structure is hybridized with yet another oligo sequence which can attach to Dynabeads®through appropriate click-chemistry