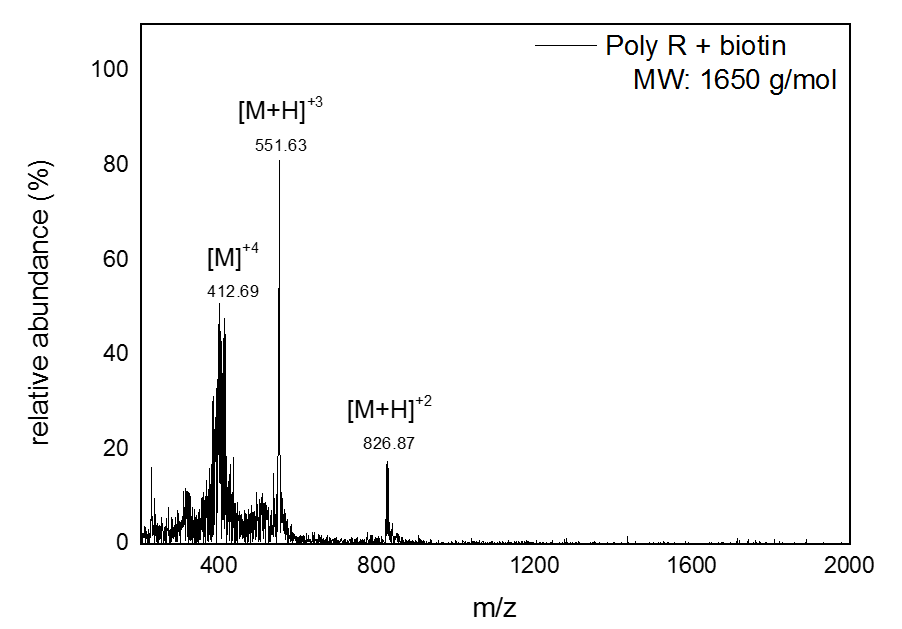
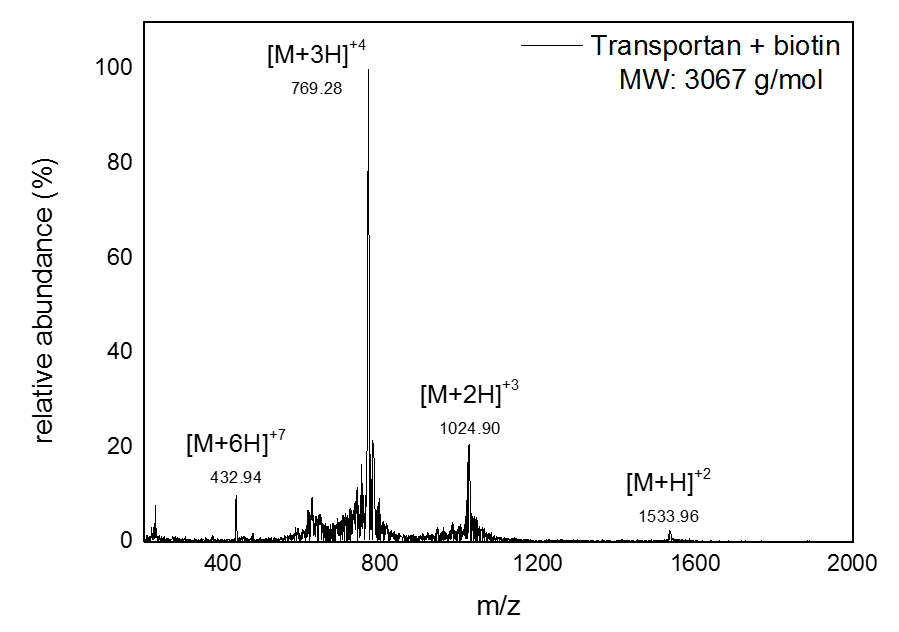
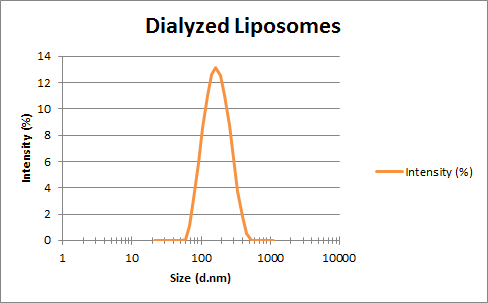
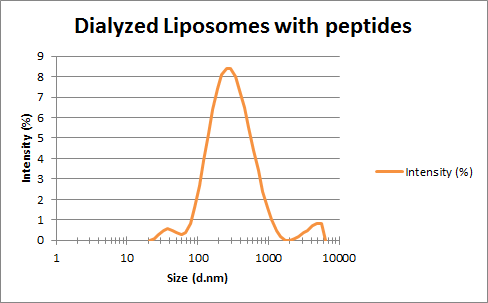
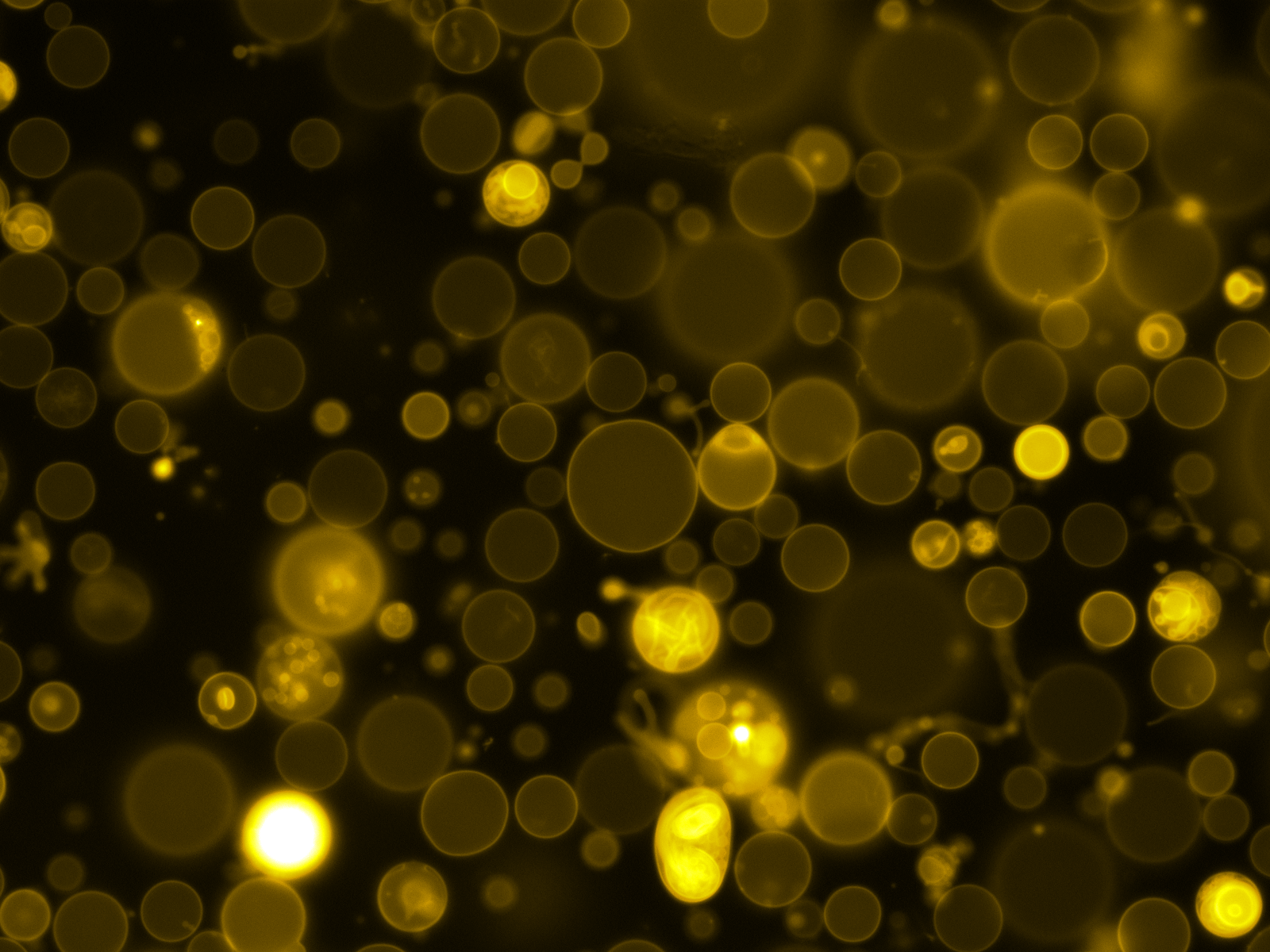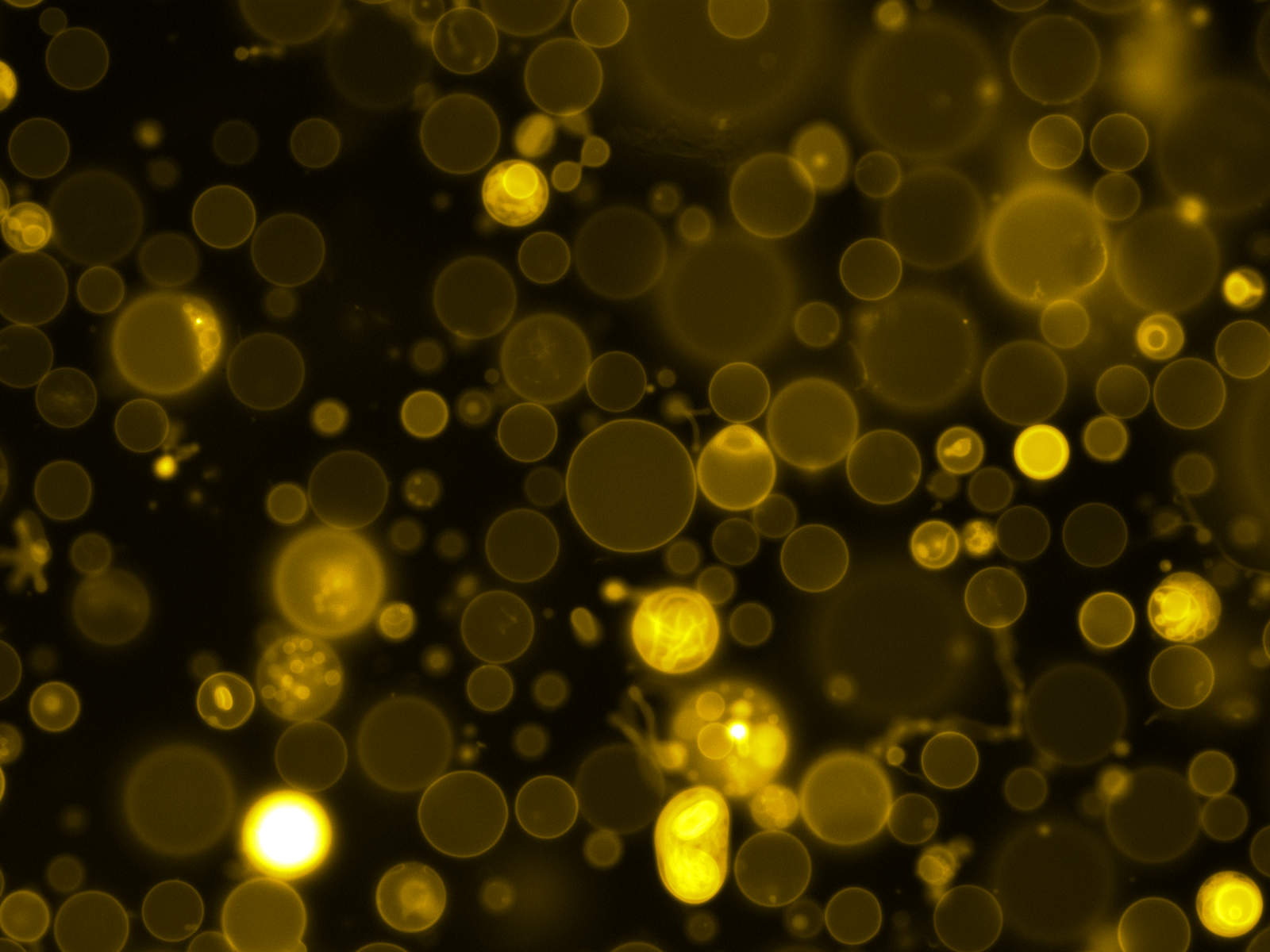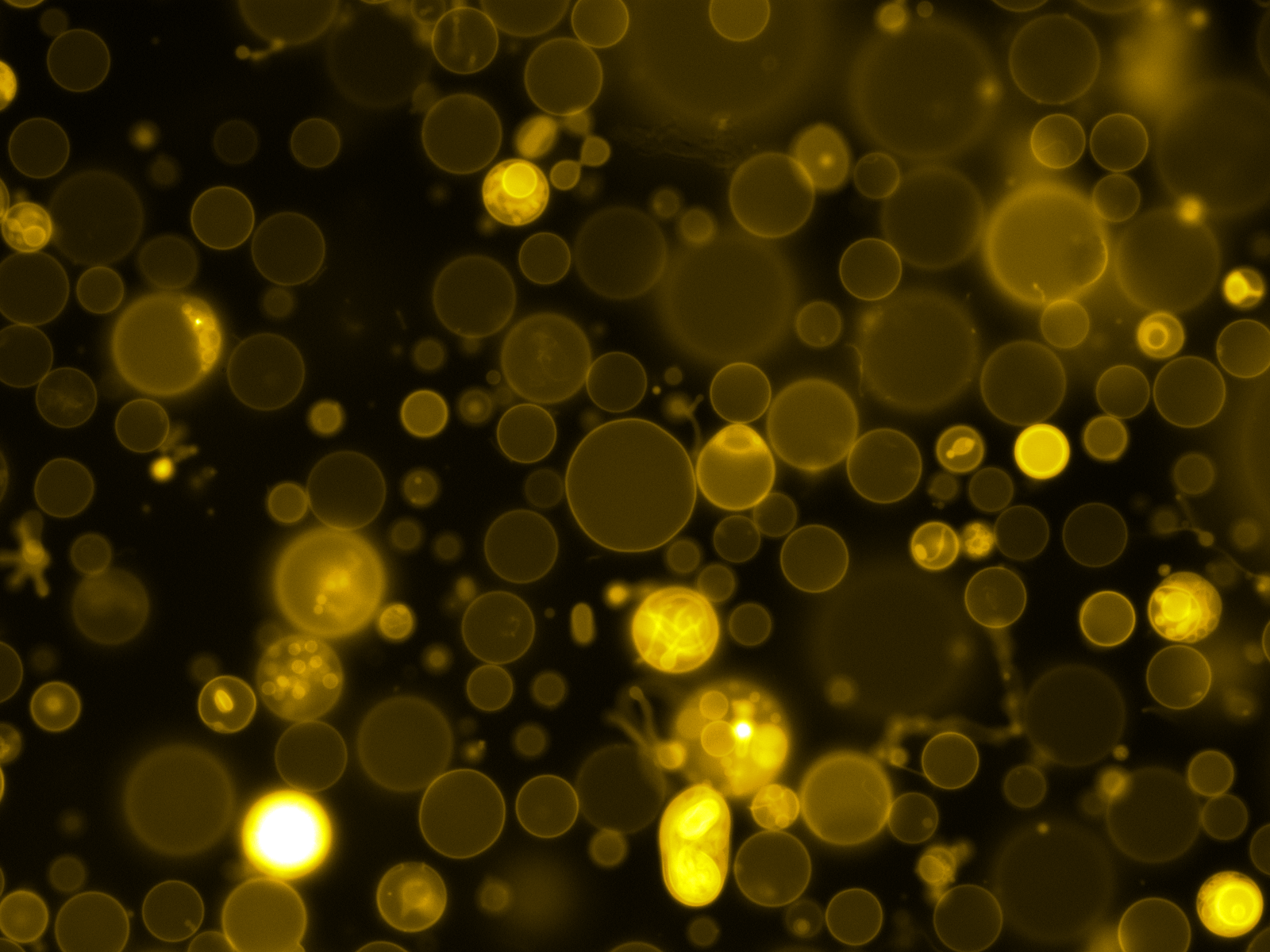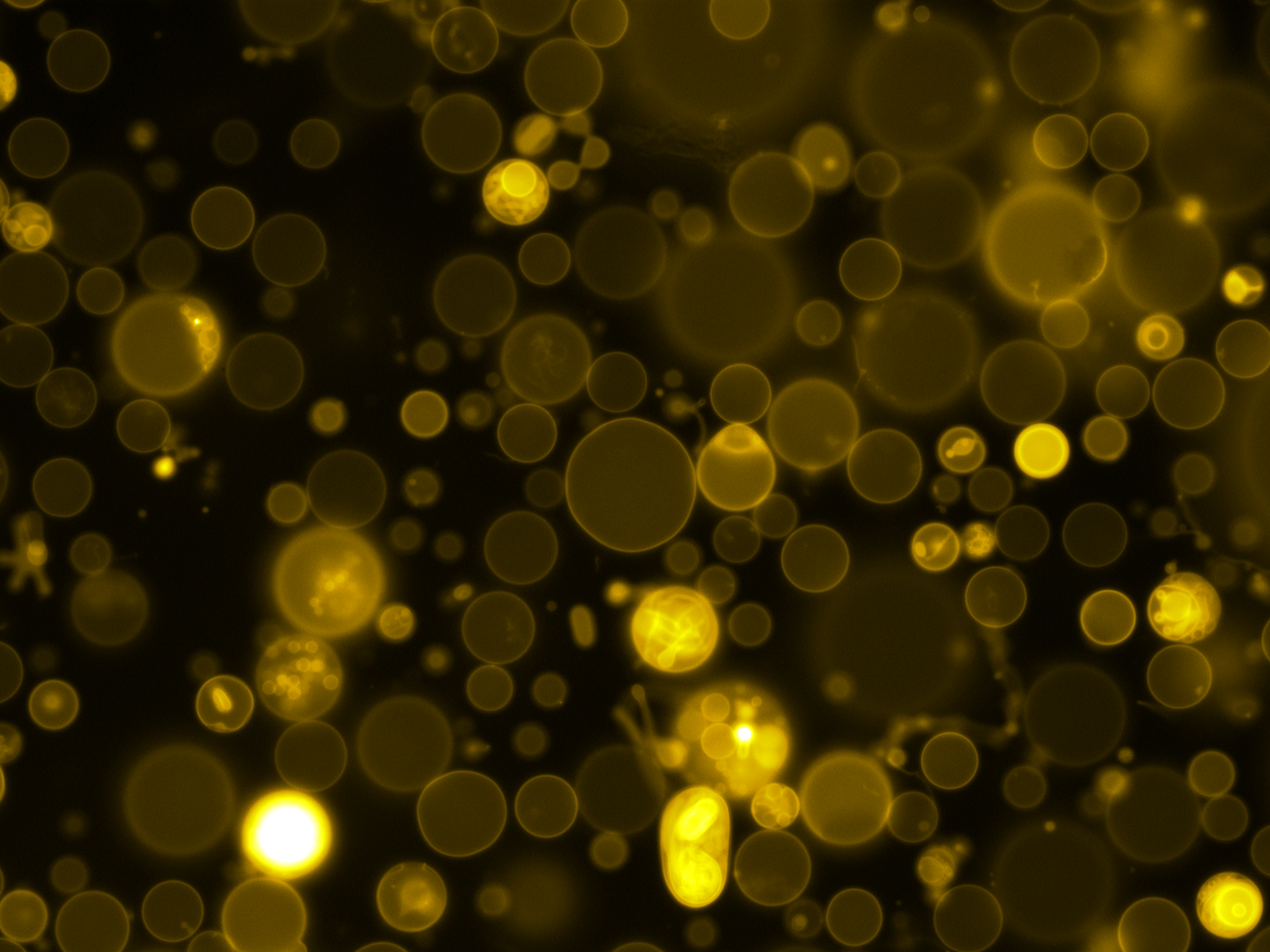
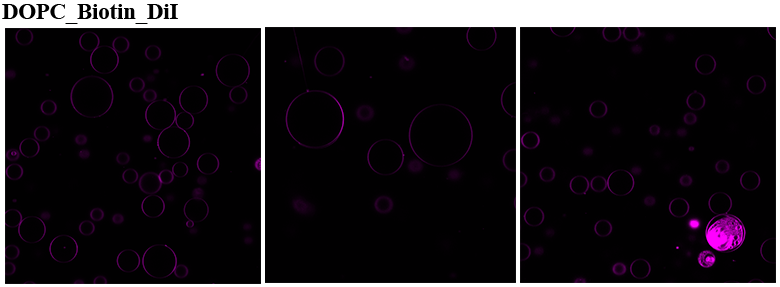
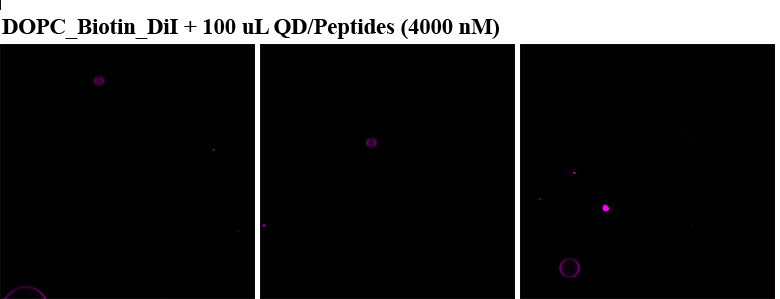
Mass Spectrometry of peptide: PolyR
Protocol
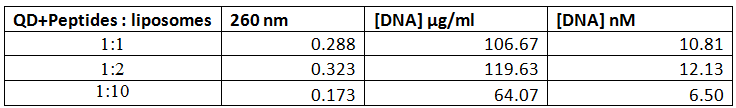
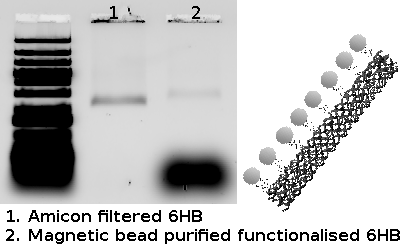
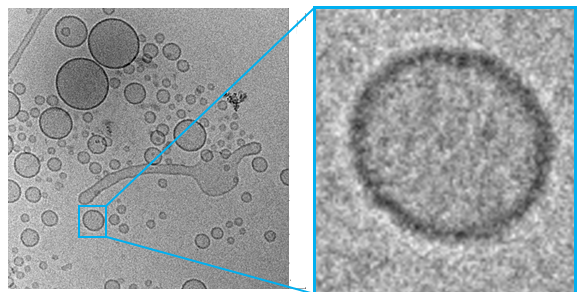
The results seen above shows the ESI-MS spectrum of conjugated Biotin to via PolyR. Since molecular ions are formed in the sample volume after being ionized through the ESI-MS, this helps researchers predict where their theoretical mass/charge ratio based on the relative abundancy as viewed in peaks. Given that the total Molecular Weight for conjugated Biotin to PolyR is 1650 g/mol, it can be safely assumed that the peak will be viewed 825 m/z. We can visibly observe this assumption to hold true, which indicate our sequence specificity was correct and successful. Other peaks that shows less mass than the molecular ion are the result of fragmentation of the molecule. This makes sense since not all solid phase peptides are properly synthesized.